Cytogenetics
Cytogenetic translocations associated with specific molecular genetic abnormalities in ALL
|
Cytogenetic |
Molecular |
% |
| cryptic t(12;21) | TEL-AML1 fusion | 25.4% |
| t(1;19)(q23;p13) | E2A-PBX (PBX1) fusion | 4.8% |
| t(9;22)(q34;q11) | BCR-ABL fusion(P185) | 1.6% |
| t(4;11)(q21;q23) | MLL-AF4Â fusion | 1.6% |
| t(8;14)(q24;q32) | IGH-MYCÂ fusion | |
| t(11;14)(p13;q11) | TCR-RBTN2 fusion |
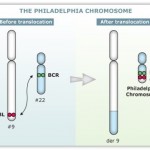 12:21 is the most common translocation and portends a good prognosis. 4:11 is the most common in children under 12 months and portends a poor prognosis.
12:21 is the most common translocation and portends a good prognosis. 4:11 is the most common in children under 12 months and portends a poor prognosis.
Prognosis
The survival rate has improved from zero four decades ago, to 20-75 percent currently, largely due to clinical trials on new chemotherapeutic agents and improvements in stem cell transplantation (SCT) technology.
Five-year survival rates evaluate older, not current, treatments. New drugs, and matching treatment to the genetic characteristics of the blast cells, may improve those rates. The prognosis for ALL differs between individuals depending on a variety of factors:
- Sex: females tend to fare better than males.
- Ethnicity: Caucasians are more likely to develop acute leukemia than African-Americans, Asians or Hispanics. However, they also tend to have a better prognosis than non-Caucasians.
- Age at diagnosis: children between 1–10 years of age are most likely to develop ALL and to be cured of it. Cases in older patients are more likely to result from chromosomal abnormalities (e.g. the Philadelphia chromosome) that make treatment more difficult and prognoses poorer.
- White blood cell count at diagnosis of less than 50,000/µl
- Cancer spread into the Central nervous system (brain or spinal cord) has worse outcomes.
- Morphological, immunological, and genetic subtypes
- Patient’s response to initial treatment
- Genetic disorders such as Down’s Syndrome
Cytogenetics, the study of characteristic large changes in the chromosomes of cancer cells, is an important predictor of outcome.
Some cytogenetic subtypes have a worse prognosis than others. These include:
- A translocation between chromosomes 9 and 22, known as the Philadelphia chromosome, occurs in about 20% of adult and 5% in pediatric cases of ALL.
- A translocation between chromosomes 4 and 11 occurs in about 4% of cases and is most common in infants under 12 months.
- Not all translocations of chromosomes carry a poorer prognosis. Some translocations are relatively favorable. For example, Hyperdiploidy (>50 chromosomes) is a good prognostic factor.
- Genome-wide copy number changes can be assessed by conventional cytogenetics or virtual karyotyping. SNP array virtual
karyotyping can detect copy number changes and LOH status, while array CGH can detect only copy number changes. Copy neutral LOH (acquired uniparental disomy) has been reported at key loci in ALL, such as CDKN2A gene, which have
prognostic significance. SNP array virtual karyotyping can readily detect copy neutral LOH. Array CGH, FISH, and conventional cytogenetics cannot detect copy neutral LOH.
|
Cytogenetic change |
Risk category |
| Philadelphia chromosome | Poor prognosis |
| t(4;11)(q21;q23) | Poor prognosis |
| t(8;14)(q24.1;q32) | Poor prognosis |
| Complex karyotype (more than four abnormalities) | Poor prognosis |
| Low hypodiploidy or near triploidy | Poor prognosis |
| High hyperdiploidy (specifically, trisomy 4, 10, 17) | Good prognosis |
| del (9p) | Good prognosis |
Correlation of prognosis with bone marrow cytogenetic finding in acute lymphoblastic leukemia
|
Prognosis |
Cytogenetic findings |
| Favorable | Hyperdiploidy > 50 ; t (12;21) |
| Intermediate | Hyperdiploidy 47 -50; Normal(diploidy); del (6q); Rearrangements of 8q24 |
| Unfavorable | Hypodiploidy-near haploidy; Near tetraploidy; del (17p); t (9;22); t (11q23) |
Unclassified ALL is considered to have an intermediate prognosis.
Classification
As ALL is not a solid tumour, the TNM notation as used in solid cancers is of little use.
The FAB classification
Subtyping of the various forms of ALL used to be done according to the French-American-British (FAB) classification, which was used for all acute leukemias (including acute myelogenous leukemia, AML).
- ALL-L1: small uniform cells
- ALL-L2: large varied cells
- ALL-L3: large varied cells with vacuoles (bubble-like features)
Each subtype is then further classified by determining the surface markers of the abnormal lymphocytes, called immunophenotyping. There are 2 main immunologic types: pre-B cell and pre-T cell. The mature B-cell ALL (L3) is now classified as Burkitt’s lymphoma/leukemia. Subtyping helps determine the prognosis and most appropriate treatment in
treating ALL.
WHO proposed classification of acute lymphoblastic leukemia
The recent WHO International panel on ALL recommends that the FAB classification be abandoned, since the morphological classification has no clinical or prognostic relevance. It instead advocates the use of the immunophenotypic classification mentioned below.
1- Acute lymphoblastic leukemia/lymphoma Synonyms:Former Fab L1/L2
- i. Precursor B acute lymphoblastic leukemia/lymphoma.
Cytogenetic subtypes:- t(12;21)(p12,q22) TEL/AML-1
- t(1;19)(q23;p13) PBX/E2A
- t(9;22)(q34;q11) ABL/BCR
- T(V,11)(V;q23) V/MLL
- ii. Precursor T acute lymphoblastic leukemia/lymphoma
2- Burkitt’s leukemia/lymphoma
Synonyms:Former FAB L3
3- Biphenotypic acute leukemia
 Variant Features of ALL
Variant Features of ALL
- 1- Acute lymphoblastic leukemia with cytoplasmic granules
- 2- Aplastic presentation of ALL
- 3- Acute lymphoblastic leukemia with eosinophilia
- 4- Relapse of lymphoblastic leukemia
- 5- Secondary ALL
Immunophenotyping in the diagnosis and classification of ALL
The use of a TdT assay and a panel of monoclonal antibodies (MoAbs) to T cell and B cell associated antigens will identify almost all cases of ALL.
Immunophenotypic categories of acute lymphoblastic leukemia (ALL)
|
Types |
FAB Class |
Tdt |
T cell associate antigen |
B cell associate antigen |
c Ig |
s Ig |
| Precursor B | L1,L2 | + | – | + | -/+ | – |
| Precursor T | L1,L2 | + | + | – | – | – |
| B-cell | L3 | – | – | + | – | + |

